Feature Selection with Boruta Part 2: Model Building
Introduction
In Part 1, we prepared the data. Now how well does Boruta work?
As a reminder, the goal is to detect gene sequences that potentially identify whether a molecule is an aptamer, which is a class of short DNA or RNA sequences with clinical and research applications. This is a difficult problem because each feature is a three, four, or five-letter genetic sequence with no obvious prior indication as to which is important. It is also a small-\(n\) large-\(p\) problem because the data sets we analyze have many more features than observations.
We will see how well Boruta works in aptamer detection for 23 different
molecules. Because this involves the same work on every molecule’s data, we will
liberally use functions from the purrr package to make our lives easier.
Prep
First, some basic prep. Because this is part of an RStudio Project, the here package
allows us to easily identify the project directory.
library(Boruta)## Loading required package: rangerlibrary(randomForest)## randomForest 4.6-14## Type rfNews() to see new features/changes/bug fixes.##
## Attaching package: 'randomForest'## The following object is masked from 'package:ranger':
##
## importancelibrary(here)## here() starts at /Users/brandonsherman/shermstats/content/post/borutalibrary(tidyverse)## ── Attaching packages ─────────────────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──## ✔ ggplot2 3.1.0 ✔ purrr 0.3.0
## ✔ tibble 2.0.1 ✔ dplyr 0.7.8
## ✔ tidyr 0.8.3 ✔ stringr 1.4.0
## ✔ readr 1.3.1 ✔ forcats 0.3.0## ── Conflicts ────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::combine() masks randomForest::combine()
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ✖ ggplot2::margin() masks randomForest::margin()library(patchwork)
SEED <- 314
input_file <- here::here("data", "atp.rds")
kmer_df <- readRDS(input_file)
kmer_df$is_aptamer <- as.factor(kmer_df$is_aptamer)
head(kmer_df)## # A tibble: 6 x 1,347
## molecule is_aptamer sequence AAA AAC AAG AAT ACA ACC ACG
## <chr> <fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 ATP 1 TCTGTTC… 0 0 1 1 1 0 0
## 2 ATP 1 GCTGAGG… 0 0 1 1 0 1 1
## 3 ATP 1 TCTGTTG… 1 0 1 1 0 0 0
## 4 ATP 1 GGACTTC… 1 1 1 0 1 1 0
## 5 ATP 1 TCTGCGC… 0 0 1 1 1 1 0
## 6 ATP 1 GCTGAGG… 0 0 1 1 0 1 1
## # … with 1,337 more variables: ACT <dbl>, AGA <dbl>, AGC <dbl>, AGG <dbl>,
## # AGT <dbl>, ATA <dbl>, ATC <dbl>, ATG <dbl>, ATT <dbl>, CAA <dbl>,
## # CAC <dbl>, CAG <dbl>, CAT <dbl>, CCA <dbl>, CCC <dbl>, CCG <dbl>,
## # CCT <dbl>, CGA <dbl>, CGC <dbl>, CGG <dbl>, CGT <dbl>, CTA <dbl>,
## # CTC <dbl>, CTG <dbl>, CTT <dbl>, GAA <dbl>, GAC <dbl>, GAG <dbl>,
## # GAT <dbl>, GCA <dbl>, GCC <dbl>, GCG <dbl>, GCT <dbl>, GGA <dbl>,
## # GGC <dbl>, GGG <dbl>, GGT <dbl>, GTA <dbl>, GTC <dbl>, GTG <dbl>,
## # GTT <dbl>, TAA <dbl>, TAC <dbl>, TAG <dbl>, TAT <dbl>, TCA <dbl>,
## # TCC <dbl>, TCG <dbl>, TCT <dbl>, TGA <dbl>, TGC <dbl>, TGG <dbl>,
## # TGT <dbl>, TTA <dbl>, TTC <dbl>, TTG <dbl>, TTT <dbl>, AAAA <dbl>,
## # AAAC <dbl>, AAAG <dbl>, AAAT <dbl>, AACA <dbl>, AACC <dbl>,
## # AACG <dbl>, AACT <dbl>, AAGA <dbl>, AAGC <dbl>, AAGG <dbl>,
## # AAGT <dbl>, AATA <dbl>, AATC <dbl>, AATG <dbl>, AATT <dbl>,
## # ACAA <dbl>, ACAC <dbl>, ACAG <dbl>, ACAT <dbl>, ACCA <dbl>,
## # ACCC <dbl>, ACCG <dbl>, ACCT <dbl>, ACGA <dbl>, ACGC <dbl>,
## # ACGG <dbl>, ACGT <dbl>, ACTA <dbl>, ACTC <dbl>, ACTG <dbl>,
## # ACTT <dbl>, AGAA <dbl>, AGAC <dbl>, AGAG <dbl>, AGAT <dbl>,
## # AGCA <dbl>, AGCC <dbl>, AGCG <dbl>, AGCT <dbl>, AGGA <dbl>,
## # AGGC <dbl>, AGGG <dbl>, …Modeling
We will follow the Boruta authors’ guide and do the following modeling for each molecule:
- Fit a random forest on the full set of 1344 predictors.
- Run Boruta.
- Fit a new random forest on the reduced set of predictors identified by Boruta.
- Compare out-of-bag error across the two models.
Build Models
Initial Random Forest and Boruta
First we define our model functions to be applied to every dataset. aptamer_random_forest will
build a random forest on a particular aptamer dataset, where the response is whether the sequence
is an aptamer, and the predictors are the presence and absence of a given k-mer. aptamer_boruta is
similar, but it runs Boruta instead.
aptamer_random_forest <- function(dat) {
## Explicitly specify X and y to avoid scoping
## issues with formulas
X <- dat %>% select(-is_aptamer)
y <- dat$is_aptamer
randomForest(X, y)
}
aptamer_boruta <- function(dat) {
X <- dat %>% select(-is_aptamer)
y <- dat$is_aptamer
Boruta(X, y, doTrace = 1)
}Now let’s build the models. Every output we wish to keep here will be appended as a column
via purrr::map. map takes in data and a function, and returns an output for every
observation. Its siblings, like map_int and map_dbl, do the same thing but return an int
or a double, respectively (and if they can’t, return an error).
The below block of code nests each molecule’s data into a list column, runs a separate random forest model (with default parameters) on each nested data.frame, and then runs a Boruta model on the same data. I have intentionally included the Boruta output because it provides a play-by-play of what the algorithm is doing. But because there’s a lot of it, I suggest looking over some of it and clicking this link to be taken to the next section.
## message=FALSE to avoid excessive output when compiled, but it will
## show up if you run the code in this chunk individually.
set.seed(SEED)
model_df <- kmer_df %>%
select(-sequence) %>%
nest(-molecule) %>%
mutate(rf_orig_fit = map(data, aptamer_random_forest))
set.seed(SEED)
model_df <- model_df %>% ## Separate in case randomization is different for the Boruta fit
mutate(boruta_fit = map(data, aptamer_boruta))## After 18 iterations, +23 secs:## confirmed 6 attributes: AATT, AATTG, ATTGC, CTGAG, GAAGA and 1 more;## rejected 1233 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1228 more;## still have 105 attributes left.## After 22 iterations, +24 secs:## confirmed 4 attributes: GAGAT, GGAAG, GTAAT, TAAT;## rejected 17 attributes: AGACA, AGACC, AGTG, ATT, CAGGA and 12 more;## still have 84 attributes left.## After 26 iterations, +24 secs:## confirmed 3 attributes: ATTGA, CTGC, GCTGA;## rejected 5 attributes: CCGTC, CGTA, GTCCG, TAT, TTGA;## still have 76 attributes left.## After 30 iterations, +25 secs:## rejected 1 attribute: GTGCG;## still have 75 attributes left.## After 33 iterations, +25 secs:## confirmed 4 attributes: AAGA, ATTG, CCGT, TTGAG;## rejected 6 attributes: AAT, AGATT, AGGCT, CTCTT, GCTCT and 1 more;## still have 65 attributes left.## After 36 iterations, +25 secs:## confirmed 3 attributes: AAGAC, AGGGC, TGAGA;## rejected 2 attributes: CCG, TTGC;## still have 60 attributes left.## After 39 iterations, +25 secs:## confirmed 1 attribute: TGAGT;## still have 59 attributes left.## After 43 iterations, +26 secs:## rejected 2 attributes: CGTCC, GATT;## still have 57 attributes left.## After 49 iterations, +26 secs:## confirmed 1 attribute: AGA;## rejected 5 attributes: AGATC, GAGTG, GCGG, TAA, TGGG;## still have 51 attributes left.## After 51 iterations, +26 secs:## confirmed 2 attributes: CAT, CGTAA;## still have 49 attributes left.## After 54 iterations, +27 secs:## rejected 1 attribute: TTGCC;## still have 48 attributes left.## After 63 iterations, +27 secs:## confirmed 1 attribute: CAACT;## still have 47 attributes left.## After 66 iterations, +28 secs:## rejected 2 attributes: CCGGG, CCGTA;## still have 45 attributes left.## After 68 iterations, +28 secs:## confirmed 1 attribute: GATTG;## still have 44 attributes left.## After 74 iterations, +28 secs:## confirmed 1 attribute: GCGGC;## still have 43 attributes left.## After 76 iterations, +28 secs:## confirmed 1 attribute: CTGCT;## still have 42 attributes left.## After 82 iterations, +29 secs:## confirmed 1 attribute: GCCGT;## still have 41 attributes left.## After 92 iterations, +30 secs:## confirmed 3 attributes: ATC, ATG, GGAA;## still have 38 attributes left.## After 95 iterations, +30 secs:## confirmed 1 attribute: GAGGG;## still have 37 attributes left.## After 97 iterations, +30 secs:## confirmed 2 attributes: GAAG, TGAG;## still have 35 attributes left.## After 18 iterations, +18 secs:## confirmed 5 attributes: GCGG, GCGGT, GGTTG, GTTGA, TTGA;## rejected 1307 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1302 more;## still have 32 attributes left.## After 22 iterations, +18 secs:## confirmed 1 attribute: GTTG;## rejected 2 attributes: TAA, TACT;## still have 29 attributes left.## After 26 iterations, +19 secs:## rejected 4 attributes: CGG, GCTA, GTT, TCGAA;## still have 25 attributes left.## After 33 iterations, +19 secs:## confirmed 1 attribute: TGA;## still have 24 attributes left.## After 36 iterations, +19 secs:## confirmed 2 attributes: CAAT, CGGT;## rejected 1 attribute: CGCGG;## still have 21 attributes left.## After 46 iterations, +19 secs:## confirmed 1 attribute: CGGTT;## still have 20 attributes left.## After 51 iterations, +19 secs:## confirmed 1 attribute: GTAGA;## still have 19 attributes left.## After 63 iterations, +20 secs:## confirmed 1 attribute: TCGA;## still have 18 attributes left.## After 66 iterations, +20 secs:## confirmed 2 attributes: GTCGA, TGAC;## still have 16 attributes left.## After 68 iterations, +20 secs:## confirmed 1 attribute: TTG;## still have 15 attributes left.## After 71 iterations, +20 secs:## confirmed 1 attribute: TCGG;## still have 14 attributes left.## After 82 iterations, +20 secs:## confirmed 2 attributes: TAGA, TTGAC;## still have 12 attributes left.## After 95 iterations, +21 secs:## rejected 1 attribute: GGA;## still have 11 attributes left.## After 18 iterations, +20 secs:## confirmed 3 attributes: AAGGG, AGGG, TGTT;## rejected 1300 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1295 more;## still have 41 attributes left.## After 22 iterations, +21 secs:## rejected 9 attributes: AAAGG, ACA, ACAA, AGGTG, ATGAG and 4 more;## still have 32 attributes left.## After 26 iterations, +21 secs:## rejected 1 attribute: AGG;## still have 31 attributes left.## After 39 iterations, +22 secs:## confirmed 2 attributes: AGGGT, AGGT;## still have 29 attributes left.## After 60 iterations, +22 secs:## confirmed 1 attribute: AGAT;## rejected 2 attributes: GAGGA, GTGCT;## still have 26 attributes left.## After 63 iterations, +23 secs:## rejected 1 attribute: TGGAA;## still have 25 attributes left.## After 68 iterations, +23 secs:## confirmed 1 attribute: ATTAG;## still have 24 attributes left.## After 74 iterations, +23 secs:## confirmed 1 attribute: GTC;## still have 23 attributes left.## After 90 iterations, +24 secs:## confirmed 1 attribute: GGGT;## still have 22 attributes left.## After 18 iterations, +22 secs:## confirmed 1 attribute: CAGC;## rejected 1314 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1309 more;## still have 29 attributes left.## After 22 iterations, +22 secs:## rejected 3 attributes: CGT, GTGTG, TTTCG;## still have 26 attributes left.## After 30 iterations, +22 secs:## confirmed 1 attribute: CAGCA;## still have 25 attributes left.## After 39 iterations, +23 secs:## confirmed 1 attribute: GGGTA;## still have 24 attributes left.## After 43 iterations, +23 secs:## confirmed 2 attributes: AGAGT, GGTC;## still have 22 attributes left.## After 49 iterations, +23 secs:## confirmed 1 attribute: AGCAC;## rejected 1 attribute: TGTGC;## still have 20 attributes left.## After 51 iterations, +23 secs:## confirmed 1 attribute: CGTGT;## still have 19 attributes left.## After 60 iterations, +24 secs:## confirmed 1 attribute: TAGTG;## still have 18 attributes left.## After 68 iterations, +24 secs:## confirmed 2 attributes: AGGGT, GAGA;## still have 16 attributes left.## After 74 iterations, +24 secs:## confirmed 1 attribute: TCAA;## still have 15 attributes left.## After 76 iterations, +24 secs:## confirmed 1 attribute: ACATA;## still have 14 attributes left.## After 79 iterations, +25 secs:## confirmed 1 attribute: CCGCG;## still have 13 attributes left.## After 82 iterations, +25 secs:## confirmed 1 attribute: AGCTA;## still have 12 attributes left.## After 84 iterations, +25 secs:## confirmed 1 attribute: GAGAC;## still have 11 attributes left.## After 90 iterations, +25 secs:## confirmed 1 attribute: GAGGC;## still have 10 attributes left.## After 95 iterations, +25 secs:## confirmed 1 attribute: GGGTC;## still have 9 attributes left.## After 18 iterations, +23 secs:## rejected 1309 attributes: AAA, AAAAA, AAAAC, AAAAG, AAAAT and 1304 more;## still have 35 attributes left.## After 22 iterations, +23 secs:## confirmed 1 attribute: GTGTG;## rejected 6 attributes: ACGTT, AGAC, ATGC, CTCG, GGTGT and 1 more;## still have 28 attributes left.## After 26 iterations, +24 secs:## confirmed 1 attribute: ATGCT;## rejected 1 attribute: CATC;## still have 26 attributes left.## After 30 iterations, +24 secs:## rejected 1 attribute: AGC;## still have 25 attributes left.## After 39 iterations, +24 secs:## rejected 1 attribute: ATCTC;## still have 24 attributes left.## After 43 iterations, +24 secs:## confirmed 1 attribute: CATGC;## still have 23 attributes left.## After 57 iterations, +25 secs:## confirmed 1 attribute: GTAAG;## still have 22 attributes left.## After 60 iterations, +25 secs:## confirmed 1 attribute: TATC;## still have 21 attributes left.## After 63 iterations, +25 secs:## confirmed 1 attribute: ATAC;## still have 20 attributes left.## After 66 iterations, +25 secs:## rejected 1 attribute: CAGG;## still have 19 attributes left.## After 68 iterations, +25 secs:## confirmed 2 attributes: AAAA, CATA;## still have 17 attributes left.## After 76 iterations, +26 secs:## confirmed 1 attribute: TCGA;## still have 16 attributes left.## After 82 iterations, +26 secs:## confirmed 1 attribute: CAC;## still have 15 attributes left.## After 84 iterations, +26 secs:## confirmed 1 attribute: GCC;## still have 14 attributes left.## After 87 iterations, +26 secs:## confirmed 1 attribute: CATG;## still have 13 attributes left.## After 18 iterations, +19 secs:## confirmed 5 attributes: AAGA, AAGAA, AGAA, CAAGA, TGCT;## rejected 1316 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1311 more;## still have 23 attributes left.## After 22 iterations, +19 secs:## confirmed 2 attributes: AGAAA, CAAG;## rejected 4 attributes: CGTTG, GAAA, GAACT, GCT;## still have 17 attributes left.## After 43 iterations, +20 secs:## confirmed 1 attribute: GATGC;## still have 16 attributes left.## After 46 iterations, +20 secs:## confirmed 1 attribute: TGCTA;## still have 15 attributes left.## After 49 iterations, +20 secs:## confirmed 1 attribute: ATGC;## still have 14 attributes left.## After 57 iterations, +20 secs:## confirmed 3 attributes: CAAGG, TACAA, TCCAA;## still have 11 attributes left.## After 60 iterations, +20 secs:## rejected 1 attribute: TGCTC;## still have 10 attributes left.## After 68 iterations, +20 secs:## confirmed 1 attribute: CTG;## still have 9 attributes left.## After 71 iterations, +21 secs:## confirmed 1 attribute: TTACA;## still have 8 attributes left.## After 74 iterations, +21 secs:## confirmed 1 attribute: CCAAG;## still have 7 attributes left.## After 84 iterations, +21 secs:## confirmed 1 attribute: GTGA;## still have 6 attributes left.## After 18 iterations, +25 secs:## confirmed 1 attribute: ACTC;## rejected 1302 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1297 more;## still have 41 attributes left.## After 22 iterations, +25 secs:## confirmed 1 attribute: TTCTC;## rejected 5 attributes: ACT, CACTG, CCTTC, GCC, TTTTT;## still have 35 attributes left.## After 30 iterations, +25 secs:## confirmed 2 attributes: ACTCC, CTTA;## rejected 1 attribute: TAA;## still have 32 attributes left.## After 36 iterations, +26 secs:## confirmed 2 attributes: CTCCC, TCCCA;## still have 30 attributes left.## After 43 iterations, +26 secs:## rejected 1 attribute: TCACC;## still have 29 attributes left.## After 46 iterations, +26 secs:## confirmed 1 attribute: GCAG;## still have 28 attributes left.## After 54 iterations, +27 secs:## confirmed 2 attributes: TCTC, TCTCC;## still have 26 attributes left.## After 57 iterations, +27 secs:## confirmed 2 attributes: CTCC, TCCA;## still have 24 attributes left.## After 60 iterations, +27 secs:## rejected 1 attribute: TCCT;## still have 23 attributes left.## After 71 iterations, +27 secs:## rejected 2 attributes: CAG, GATC;## still have 21 attributes left.## After 74 iterations, +27 secs:## confirmed 1 attribute: CTCTC;## still have 20 attributes left.## After 76 iterations, +28 secs:## confirmed 1 attribute: TCTCT;## still have 19 attributes left.## After 84 iterations, +28 secs:## confirmed 1 attribute: GGATA;## still have 18 attributes left.## After 95 iterations, +28 secs:## rejected 1 attribute: ACCC;## still have 17 attributes left.## After 18 iterations, +14 secs:## confirmed 1 attribute: GGGA;## rejected 1322 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1317 more;## still have 21 attributes left.## After 22 iterations, +14 secs:## rejected 2 attributes: CGTAG, GGGAT;## still have 19 attributes left.## After 60 iterations, +15 secs:## rejected 1 attribute: AGGG;## still have 18 attributes left.## After 76 iterations, +16 secs:## confirmed 1 attribute: GCGT;## still have 17 attributes left.## After 92 iterations, +16 secs:## confirmed 1 attribute: AAGA;## still have 16 attributes left.## After 95 iterations, +17 secs:## confirmed 1 attribute: GGCGT;## still have 15 attributes left.## After 18 iterations, +23 secs:## confirmed 5 attributes: AATCA, CAT, TCA, TCAC, TCAT;## rejected 1294 attributes: AAAA, AAAAA, AAAAC, AAAAG, AAAAT and 1289 more;## still have 45 attributes left.## After 22 iterations, +23 secs:## confirmed 5 attributes: ACTCA, ATCA, ATCAC, GATTC, GTCAT;## rejected 6 attributes: AAC, ACTAT, TATG, TGAGT, TTT and 1 more;## still have 34 attributes left.## After 26 iterations, +23 secs:## confirmed 1 attribute: TGATT;## rejected 2 attributes: CAC, TCATC;## still have 31 attributes left.## After 30 iterations, +24 secs:## confirmed 2 attributes: ATGA, TCATA;## rejected 4 attributes: AAA, CACT, TCGA, TTCAT;## still have 25 attributes left.## After 33 iterations, +24 secs:## confirmed 1 attribute: TCACT;## still have 24 attributes left.## After 36 iterations, +24 secs:## rejected 1 attribute: AGTA;## still have 23 attributes left.## After 39 iterations, +24 secs:## confirmed 1 attribute: TACTC;## still have 22 attributes left.## After 43 iterations, +24 secs:## confirmed 1 attribute: AGTCA;## still have 21 attributes left.## After 51 iterations, +24 secs:## confirmed 1 attribute: TGAT;## still have 20 attributes left.## After 54 iterations, +25 secs:## confirmed 2 attributes: ATTC, CTCA;## still have 18 attributes left.## After 57 iterations, +25 secs:## confirmed 1 attribute: AATC;## still have 17 attributes left.## After 63 iterations, +25 secs:## rejected 1 attribute: ACAT;## still have 16 attributes left.## After 76 iterations, +25 secs:## confirmed 1 attribute: TTACT;## still have 15 attributes left.## After 97 iterations, +26 secs:## rejected 1 attribute: ATCAT;## still have 14 attributes left.## After 18 iterations, +1.2 mins:## confirmed 34 attributes: ACG, ACGC, ACGT, ACGTA, ATT and 29 more;## rejected 1173 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1168 more;## still have 137 attributes left.## After 22 iterations, +1.2 mins:## confirmed 4 attributes: CCTGG, CGTCG, GCACG, TGG;## rejected 9 attributes: AATGC, ATCAC, CTC, CTGTT, TACGG and 4 more;## still have 124 attributes left.## After 26 iterations, +1.2 mins:## confirmed 7 attributes: AAGTT, AATG, ACGCA, AGTTG, GCATG and 2 more;## rejected 5 attributes: CCGG, GAGG, GGGA, TAG, TGTA;## still have 112 attributes left.## After 30 iterations, +1.3 mins:## confirmed 2 attributes: CATGC, TAAT;## rejected 3 attributes: CAGCA, GGGAT, GTTC;## still have 107 attributes left.## After 33 iterations, +1.3 mins:## confirmed 2 attributes: GGGCC, GTGGG;## rejected 7 attributes: AAGC, AAGT, ACGA, ACGAG, CTG and 2 more;## still have 98 attributes left.## After 36 iterations, +1.3 mins:## confirmed 3 attributes: GAAA, GAAAG, TCCGG;## still have 95 attributes left.## After 39 iterations, +1.3 mins:## rejected 1 attribute: GTTGG;## still have 94 attributes left.## After 43 iterations, +1.3 mins:## confirmed 7 attributes: AGTGG, CACGC, CATCT, CCGTA, GCCTG and 2 more;## still have 87 attributes left.## After 46 iterations, +1.4 mins:## confirmed 4 attributes: AGGAG, CGTAT, TCG, TGCCT;## still have 83 attributes left.## After 49 iterations, +1.4 mins:## confirmed 1 attribute: AGCGT;## rejected 4 attributes: AGGT, ATGA, CCT, CGAC;## still have 78 attributes left.## After 51 iterations, +1.4 mins:## confirmed 2 attributes: GTAA, TACGC;## rejected 2 attributes: AAC, TCT;## still have 74 attributes left.## After 54 iterations, +1.4 mins:## confirmed 1 attribute: ACCGT;## still have 73 attributes left.## After 57 iterations, +1.4 mins:## confirmed 3 attributes: CGGT, TTC, TTCG;## still have 70 attributes left.## After 60 iterations, +1.5 mins:## confirmed 1 attribute: AAAGT;## rejected 1 attribute: TCGG;## still have 68 attributes left.## After 63 iterations, +1.5 mins:## confirmed 1 attribute: GGAGC;## rejected 1 attribute: GCTAT;## still have 66 attributes left.## After 66 iterations, +1.5 mins:## confirmed 2 attributes: AATGA, CGACC;## still have 64 attributes left.## After 74 iterations, +1.6 mins:## confirmed 2 attributes: AGT, TAGT;## still have 62 attributes left.## After 76 iterations, +1.6 mins:## confirmed 6 attributes: AGTT, CACAG, CGCAT, GCAT, GCCT and 1 more;## rejected 1 attribute: TGGGG;## still have 55 attributes left.## After 82 iterations, +1.6 mins:## rejected 1 attribute: CAGG;## still have 54 attributes left.## After 84 iterations, +1.6 mins:## rejected 1 attribute: TTCT;## still have 53 attributes left.## After 87 iterations, +1.7 mins:## confirmed 2 attributes: AAG, GGTA;## still have 51 attributes left.## After 90 iterations, +1.7 mins:## confirmed 2 attributes: ATGCC, GGT;## still have 49 attributes left.## After 92 iterations, +1.7 mins:## confirmed 1 attribute: ATG;## still have 48 attributes left.## After 95 iterations, +1.7 mins:## rejected 1 attribute: CACTG;## still have 47 attributes left.## After 97 iterations, +1.7 mins:## confirmed 1 attribute: GTACG;## still have 46 attributes left.## After 18 iterations, +21 secs:## rejected 1328 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1323 more;## still have 16 attributes left.## After 22 iterations, +21 secs:## confirmed 1 attribute: CTCTC;## still have 15 attributes left.## After 26 iterations, +21 secs:## rejected 1 attribute: TATC;## still have 14 attributes left.## After 30 iterations, +21 secs:## rejected 1 attribute: GTG;## still have 13 attributes left.## After 43 iterations, +22 secs:## confirmed 1 attribute: CCGT;## still have 12 attributes left.## After 51 iterations, +22 secs:## confirmed 1 attribute: ATCC;## still have 11 attributes left.## After 60 iterations, +22 secs:## rejected 1 attribute: AGCT;## still have 10 attributes left.## After 63 iterations, +22 secs:## rejected 1 attribute: CCCG;## still have 9 attributes left.## After 71 iterations, +22 secs:## rejected 1 attribute: CCTCT;## still have 8 attributes left.## After 79 iterations, +23 secs:## confirmed 1 attribute: TGTG;## still have 7 attributes left.## After 84 iterations, +23 secs:## confirmed 1 attribute: GGG;## still have 6 attributes left.## After 18 iterations, +15 secs:## confirmed 11 attributes: AAACT, AACT, AACTC, AAGA, AGAA and 6 more;## rejected 1259 attributes: AAAA, AAAAA, AAAAC, AAAAT, AAACA and 1254 more;## still have 74 attributes left.## After 22 iterations, +16 secs:## confirmed 1 attribute: ACTC;## rejected 15 attributes: ACTGT, AGA, ATA, CACT, CCCAG and 10 more;## still have 58 attributes left.## After 26 iterations, +16 secs:## confirmed 2 attributes: ACTTC, GCAC;## rejected 2 attributes: ACT, CAAAA;## still have 54 attributes left.## After 30 iterations, +16 secs:## confirmed 1 attribute: TGGC;## rejected 3 attributes: CAT, GCACT, GTTG;## still have 50 attributes left.## After 33 iterations, +16 secs:## confirmed 1 attribute: TCGG;## rejected 5 attributes: CAGG, CGA, CTGTG, GCCA, TGTG;## still have 44 attributes left.## After 36 iterations, +17 secs:## confirmed 1 attribute: GGAA;## still have 43 attributes left.## After 39 iterations, +17 secs:## confirmed 1 attribute: AAGAC;## rejected 1 attribute: CTGT;## still have 41 attributes left.## After 43 iterations, +17 secs:## confirmed 2 attributes: CGGTG, GGAC;## rejected 2 attributes: AGCAA, CGT;## still have 37 attributes left.## After 49 iterations, +17 secs:## rejected 1 attribute: AAAAG;## still have 36 attributes left.## After 54 iterations, +17 secs:## confirmed 1 attribute: TTC;## rejected 1 attribute: GAAA;## still have 34 attributes left.## After 60 iterations, +18 secs:## confirmed 3 attributes: GGCAC, TCGGT, TGTGG;## still have 31 attributes left.## After 74 iterations, +18 secs:## confirmed 1 attribute: AAAC;## still have 30 attributes left.## After 76 iterations, +18 secs:## confirmed 2 attributes: TTCG, TTCGG;## rejected 1 attribute: GGT;## still have 27 attributes left.## After 84 iterations, +19 secs:## rejected 1 attribute: ACTCG;## still have 26 attributes left.## After 92 iterations, +19 secs:## confirmed 1 attribute: GGGA;## still have 25 attributes left.## After 18 iterations, +45 secs:## confirmed 4 attributes: ATCAC, GGAT, GGC, GTCTG;## rejected 1256 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1251 more;## still have 84 attributes left.## After 22 iterations, +45 secs:## confirmed 1 attribute: ATCA;## rejected 19 attributes: AAAT, AAG, AGGC, ATCG, ATCT and 14 more;## still have 64 attributes left.## After 26 iterations, +46 secs:## rejected 3 attributes: ATCAG, CTGCA, TGCAG;## still have 61 attributes left.## After 30 iterations, +46 secs:## confirmed 2 attributes: GCAG, TGCA;## rejected 3 attributes: AGTC, CAGGA, TCAAG;## still have 56 attributes left.## After 33 iterations, +47 secs:## confirmed 1 attribute: TTTC;## rejected 2 attributes: CAAGT, GGAG;## still have 53 attributes left.## After 36 iterations, +47 secs:## confirmed 2 attributes: GCTC, GGTCT;## rejected 3 attributes: ACAA, CACAG, CTGT;## still have 48 attributes left.## After 39 iterations, +47 secs:## rejected 1 attribute: CCTCT;## still have 47 attributes left.## After 49 iterations, +48 secs:## confirmed 2 attributes: GGTC, TACA;## still have 45 attributes left.## After 51 iterations, +49 secs:## confirmed 1 attribute: GATCA;## still have 44 attributes left.## After 54 iterations, +49 secs:## confirmed 1 attribute: AGT;## still have 43 attributes left.## After 57 iterations, +49 secs:## confirmed 1 attribute: TCCTC;## still have 42 attributes left.## After 60 iterations, +49 secs:## confirmed 2 attributes: GCA, GGATT;## still have 40 attributes left.## After 63 iterations, +50 secs:## confirmed 2 attributes: AACG, TCAC;## still have 38 attributes left.## After 66 iterations, +50 secs:## confirmed 1 attribute: ACGGG;## rejected 1 attribute: CTC;## still have 36 attributes left.## After 68 iterations, +50 secs:## rejected 1 attribute: CGGC;## still have 35 attributes left.## After 74 iterations, +51 secs:## confirmed 1 attribute: TTTTC;## still have 34 attributes left.## After 84 iterations, +52 secs:## rejected 1 attribute: TTGGA;## still have 33 attributes left.## After 87 iterations, +52 secs:## confirmed 1 attribute: CATCA;## still have 32 attributes left.## After 92 iterations, +53 secs:## rejected 1 attribute: AAT;## still have 31 attributes left.## After 18 iterations, +14 secs:## rejected 1338 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1333 more;## still have 6 attributes left.## After 71 iterations, +14 secs:## confirmed 1 attribute: AGGG;## still have 5 attributes left.## After 95 iterations, +15 secs:## confirmed 1 attribute: TTCTC;## still have 4 attributes left.## After 18 iterations, +13 secs:## confirmed 4 attributes: ACCCG, CCCG, CCT, CGT;## rejected 1250 attributes: AAAA, AAAAA, AAAAC, AAAAT, AAAC and 1245 more;## still have 90 attributes left.## After 22 iterations, +13 secs:## confirmed 1 attribute: ACG;## rejected 31 attributes: AAAAG, AAAG, AAAGA, ACAGA, ACTA and 26 more;## still have 58 attributes left.## After 26 iterations, +13 secs:## rejected 2 attributes: CCCT, TGGT;## still have 56 attributes left.## After 33 iterations, +13 secs:## rejected 4 attributes: AACAT, CTAC, TATGG, TTC;## still have 52 attributes left.## After 36 iterations, +14 secs:## confirmed 3 attributes: CCCGG, GCA, GCTC;## rejected 6 attributes: AAGA, CAGTT, CTGT, GCGCC, GGCGC and 1 more;## still have 43 attributes left.## After 39 iterations, +14 secs:## rejected 3 attributes: CGGCG, CTTAC, TCCG;## still have 40 attributes left.## After 43 iterations, +14 secs:## confirmed 1 attribute: CTAT;## rejected 1 attribute: ATC;## still have 38 attributes left.## After 46 iterations, +14 secs:## confirmed 2 attributes: CCGC, TACTT;## rejected 1 attribute: CTGA;## still have 35 attributes left.## After 49 iterations, +14 secs:## rejected 3 attributes: ATG, CCCTT, TACT;## still have 32 attributes left.## After 51 iterations, +14 secs:## confirmed 2 attributes: AGTTC, GCCC;## still have 30 attributes left.## After 54 iterations, +14 secs:## confirmed 1 attribute: GGAAA;## still have 29 attributes left.## After 57 iterations, +15 secs:## confirmed 1 attribute: CGCG;## rejected 1 attribute: TGGGC;## still have 27 attributes left.## After 60 iterations, +15 secs:## confirmed 1 attribute: GCCG;## still have 26 attributes left.## After 66 iterations, +15 secs:## confirmed 1 attribute: ACTCC;## still have 25 attributes left.## After 82 iterations, +16 secs:## confirmed 1 attribute: ACTGG;## still have 24 attributes left.## After 87 iterations, +16 secs:## confirmed 1 attribute: CAG;## still have 23 attributes left.## After 97 iterations, +16 secs:## rejected 1 attribute: ATGG;## still have 22 attributes left.## After 18 iterations, +1.1 mins:## confirmed 29 attributes: AACCC, AATC, AATCC, ATAA, ATTC and 24 more;## rejected 1255 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1250 more;## still have 60 attributes left.## After 22 iterations, +1.1 mins:## confirmed 2 attributes: AGC, ATA;## rejected 9 attributes: AAG, ACT, AGTTG, CCCGC, CGGGG and 4 more;## still have 49 attributes left.## After 26 iterations, +1.1 mins:## confirmed 3 attributes: AACC, GCGAT, TAA;## rejected 7 attributes: ATTGC, CCCCC, CGAT, GAACC, TAAG and 2 more;## still have 39 attributes left.## After 30 iterations, +1.1 mins:## confirmed 1 attribute: GAA;## rejected 2 attributes: CGGA, GCG;## still have 36 attributes left.## After 33 iterations, +1.2 mins:## rejected 1 attribute: AAT;## still have 35 attributes left.## After 39 iterations, +1.2 mins:## confirmed 1 attribute: GAAT;## still have 34 attributes left.## After 43 iterations, +1.2 mins:## confirmed 1 attribute: TAAA;## still have 33 attributes left.## After 46 iterations, +1.2 mins:## confirmed 1 attribute: TAGC;## rejected 1 attribute: TAAGG;## still have 31 attributes left.## After 49 iterations, +1.2 mins:## rejected 2 attributes: AAC, GAC;## still have 29 attributes left.## After 51 iterations, +1.2 mins:## confirmed 1 attribute: ACCCC;## rejected 1 attribute: TTC;## still have 27 attributes left.## After 57 iterations, +1.2 mins:## rejected 1 attribute: AATAC;## still have 26 attributes left.## After 60 iterations, +1.2 mins:## rejected 1 attribute: GGC;## still have 25 attributes left.## After 71 iterations, +1.3 mins:## rejected 1 attribute: TCCCC;## still have 24 attributes left.## After 74 iterations, +1.3 mins:## confirmed 1 attribute: GGGGG;## rejected 1 attribute: ATT;## still have 22 attributes left.## After 76 iterations, +1.3 mins:## rejected 1 attribute: TGAT;## still have 21 attributes left.## After 18 iterations, +18 secs:## confirmed 17 attributes: AAACT, AACTG, AAGAA, ACT, ACTG and 12 more;## rejected 1275 attributes: AAAA, AAAAA, AAAAC, AAAAG, AAAAT and 1270 more;## still have 52 attributes left.## After 22 iterations, +18 secs:## rejected 7 attributes: AAA, AAGG, CACTG, GAAA, GGAT and 2 more;## still have 45 attributes left.## After 26 iterations, +18 secs:## confirmed 3 attributes: AGA, AGAAA, GAAG;## rejected 3 attributes: ATA, ATG, GCAT;## still have 39 attributes left.## After 30 iterations, +18 secs:## confirmed 2 attributes: GGAA, GTGGA;## rejected 2 attributes: AGTC, CCA;## still have 35 attributes left.## After 33 iterations, +18 secs:## rejected 4 attributes: AGAAC, ATGG, GCCA, TGAT;## still have 31 attributes left.## After 36 iterations, +19 secs:## confirmed 2 attributes: GTGA, TGA;## rejected 1 attribute: ATCA;## still have 28 attributes left.## After 39 iterations, +19 secs:## confirmed 1 attribute: AAGA;## rejected 1 attribute: AGGG;## still have 26 attributes left.## After 49 iterations, +19 secs:## confirmed 2 attributes: GTAGA, TGCA;## still have 24 attributes left.## After 51 iterations, +19 secs:## rejected 1 attribute: CCTGC;## still have 23 attributes left.## After 57 iterations, +19 secs:## rejected 2 attributes: CGGA, GAG;## still have 21 attributes left.## After 68 iterations, +20 secs:## confirmed 1 attribute: AACT;## still have 20 attributes left.## After 71 iterations, +20 secs:## confirmed 1 attribute: TGG;## rejected 1 attribute: TAAG;## still have 18 attributes left.## After 76 iterations, +20 secs:## confirmed 2 attributes: GAA, GACTG;## still have 16 attributes left.## After 82 iterations, +21 secs:## confirmed 1 attribute: TGCGA;## still have 15 attributes left.## After 87 iterations, +21 secs:## confirmed 1 attribute: TAGA;## still have 14 attributes left.## After 18 iterations, +19 secs:## confirmed 17 attributes: AAGAA, AAT, AATT, AATTG, AGAAT and 12 more;## rejected 1267 attributes: AAAA, AAAAA, AAAAC, AAAAG, AAAAT and 1262 more;## still have 60 attributes left.## After 22 iterations, +20 secs:## confirmed 2 attributes: AATGC, TTATA;## rejected 10 attributes: AGA, AGTG, ATGG, CGC, CGT and 5 more;## still have 48 attributes left.## After 26 iterations, +20 secs:## confirmed 2 attributes: AAGA, AGAA;## rejected 1 attribute: TGGAT;## still have 45 attributes left.## After 30 iterations, +20 secs:## confirmed 3 attributes: CAC, CTTAT, TGAA;## rejected 1 attribute: GCGA;## still have 41 attributes left.## After 33 iterations, +20 secs:## rejected 3 attributes: GTC, TCCT, TGCT;## still have 38 attributes left.## After 36 iterations, +20 secs:## confirmed 1 attribute: CCT;## rejected 2 attributes: AGTGA, GCTT;## still have 35 attributes left.## After 39 iterations, +21 secs:## confirmed 2 attributes: AATG, ACGGA;## still have 33 attributes left.## After 46 iterations, +21 secs:## confirmed 1 attribute: CGGAA;## still have 32 attributes left.## After 49 iterations, +21 secs:## confirmed 1 attribute: CAGTG;## still have 31 attributes left.## After 54 iterations, +21 secs:## confirmed 1 attribute: CCGAT;## still have 30 attributes left.## After 60 iterations, +22 secs:## confirmed 1 attribute: GAAG;## still have 29 attributes left.## After 66 iterations, +22 secs:## confirmed 1 attribute: AAA;## still have 28 attributes left.## After 68 iterations, +22 secs:## confirmed 1 attribute: ATGC;## still have 27 attributes left.## After 76 iterations, +23 secs:## confirmed 1 attribute: ATT;## still have 26 attributes left.## After 82 iterations, +23 secs:## confirmed 1 attribute: TTG;## still have 25 attributes left.## After 84 iterations, +23 secs:## confirmed 1 attribute: AACCG;## still have 24 attributes left.## After 87 iterations, +23 secs:## confirmed 1 attribute: CACT;## rejected 1 attribute: GTCT;## still have 22 attributes left.## After 90 iterations, +23 secs:## rejected 1 attribute: ACGG;## still have 21 attributes left.## After 97 iterations, +24 secs:## confirmed 1 attribute: GTGA;## still have 20 attributes left.## After 18 iterations, +13 secs:## confirmed 2 attributes: ATG, TATG;## rejected 1308 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1303 more;## still have 34 attributes left.## After 22 iterations, +13 secs:## confirmed 4 attributes: ATGG, ATGGT, TATGG, TGG;## rejected 6 attributes: AAGG, ATGC, CGT, GCA, GCTAT and 1 more;## still have 24 attributes left.## After 26 iterations, +13 secs:## rejected 1 attribute: CAG;## still have 23 attributes left.## After 30 iterations, +13 secs:## confirmed 1 attribute: AATG;## rejected 2 attributes: AGGC, GCCCC;## still have 20 attributes left.## After 36 iterations, +14 secs:## confirmed 1 attribute: ACCC;## rejected 1 attribute: TGGTG;## still have 18 attributes left.## After 46 iterations, +14 secs:## rejected 1 attribute: CAGT;## still have 17 attributes left.## After 49 iterations, +14 secs:## rejected 1 attribute: GATA;## still have 16 attributes left.## After 54 iterations, +14 secs:## confirmed 1 attribute: CCC;## still have 15 attributes left.## After 66 iterations, +14 secs:## confirmed 1 attribute: TGCT;## rejected 1 attribute: CCCC;## still have 13 attributes left.## After 18 iterations, +14 secs:## confirmed 5 attributes: ATCC, ATCCC, GATC, GATCC, TCCC;## rejected 1314 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1309 more;## still have 25 attributes left.## After 22 iterations, +14 secs:## rejected 4 attributes: CCAA, CCAC, CGCGC, TCCG;## still have 21 attributes left.## After 33 iterations, +15 secs:## rejected 1 attribute: TCCCT;## still have 20 attributes left.## After 36 iterations, +15 secs:## rejected 1 attribute: GCA;## still have 19 attributes left.## After 49 iterations, +15 secs:## confirmed 1 attribute: CAC;## still have 18 attributes left.## After 54 iterations, +15 secs:## confirmed 1 attribute: GGAT;## still have 17 attributes left.## After 63 iterations, +15 secs:## confirmed 2 attributes: AGGGG, GGATC;## still have 15 attributes left.## After 71 iterations, +16 secs:## confirmed 1 attribute: GGCA;## still have 14 attributes left.## After 74 iterations, +16 secs:## confirmed 1 attribute: GGGCT;## still have 13 attributes left.## After 76 iterations, +16 secs:## confirmed 1 attribute: CACT;## still have 12 attributes left.## After 87 iterations, +16 secs:## confirmed 1 attribute: GGCTC;## still have 11 attributes left.## After 95 iterations, +16 secs:## confirmed 1 attribute: TCCCG;## still have 10 attributes left.## After 97 iterations, +16 secs:## confirmed 1 attribute: AGGG;## still have 9 attributes left.## After 18 iterations, +1 mins:## confirmed 4 attributes: ACA, ACAC, CAC, TACA;## rejected 1268 attributes: AAAAA, AAAAG, AAAAT, AAAC, AAACA and 1263 more;## still have 72 attributes left.## After 22 iterations, +1 mins:## confirmed 5 attributes: ACAT, CACG, CCC, TACAC, TAG;## rejected 16 attributes: AAAT, AAATG, AATC, ACAAG, ACACT and 11 more;## still have 51 attributes left.## After 26 iterations, +1 mins:## confirmed 1 attribute: GAA;## rejected 5 attributes: AAAAC, ATC, CATTG, CCCAA, GCACA;## still have 45 attributes left.## After 30 iterations, +1.1 mins:## confirmed 1 attribute: TGCA;## rejected 1 attribute: GAAC;## still have 43 attributes left.## After 33 iterations, +1.1 mins:## confirmed 1 attribute: AAG;## still have 42 attributes left.## After 36 iterations, +1.1 mins:## confirmed 3 attributes: CACAC, CACGC, TACAT;## rejected 3 attributes: AAT, TAACC, TCCC;## still have 36 attributes left.## After 39 iterations, +1.1 mins:## confirmed 1 attribute: AAAA;## still have 35 attributes left.## After 46 iterations, +1.1 mins:## rejected 1 attribute: CACAT;## still have 34 attributes left.## After 49 iterations, +1.1 mins:## confirmed 1 attribute: CAAAA;## still have 33 attributes left.## After 54 iterations, +1.1 mins:## confirmed 2 attributes: AACC, AGAA;## rejected 1 attribute: GAAA;## still have 30 attributes left.## After 60 iterations, +1.1 mins:## confirmed 2 attributes: TAGC, TCC;## still have 28 attributes left.## After 66 iterations, +1.1 mins:## confirmed 1 attribute: CCCA;## rejected 1 attribute: AAA;## still have 26 attributes left.## After 71 iterations, +1.1 mins:## confirmed 2 attributes: ATACA, CCAA;## still have 24 attributes left.## After 74 iterations, +1.1 mins:## confirmed 1 attribute: CGACC;## still have 23 attributes left.## After 76 iterations, +1.2 mins:## confirmed 2 attributes: ACAAC, TAC;## still have 21 attributes left.## After 82 iterations, +1.2 mins:## rejected 1 attribute: ACATG;## still have 20 attributes left.## After 18 iterations, +23 secs:## confirmed 3 attributes: ACCG, GTACC, TACC;## rejected 1323 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1318 more;## still have 18 attributes left.## After 22 iterations, +24 secs:## rejected 2 attributes: CCGC, GTAC;## still have 16 attributes left.## After 36 iterations, +24 secs:## confirmed 1 attribute: TACCG;## still have 15 attributes left.## After 51 iterations, +25 secs:## rejected 1 attribute: TAC;## still have 14 attributes left.## After 54 iterations, +25 secs:## confirmed 2 attributes: ACCGT, AGTAC;## still have 12 attributes left.## After 68 iterations, +25 secs:## confirmed 2 attributes: GTGTG, TGGTG;## still have 10 attributes left.## After 71 iterations, +25 secs:## rejected 1 attribute: TTC;## still have 9 attributes left.## After 79 iterations, +26 secs:## confirmed 1 attribute: GCTTA;## rejected 1 attribute: CCGT;## still have 7 attributes left.## After 82 iterations, +26 secs:## confirmed 1 attribute: GGCTT;## still have 6 attributes left.## After 18 iterations, +19 secs:## confirmed 28 attributes: AGTT, AGTTG, ATAGA, CGATG, CTGT and 23 more;## rejected 1235 attributes: AAA, AAAA, AAAAA, AAAAC, AAAAG and 1230 more;## still have 81 attributes left.## After 22 iterations, +19 secs:## confirmed 3 attributes: ACGTG, CCGAT, CGA;## rejected 20 attributes: AAG, AATGG, ACGTT, ACTAT, AGC and 15 more;## still have 58 attributes left.## After 26 iterations, +19 secs:## confirmed 3 attributes: AATGA, GCT, TGC;## rejected 5 attributes: ACGC, AGT, TCA, TCTAC, TGAAC;## still have 50 attributes left.## After 30 iterations, +19 secs:## rejected 2 attributes: CCG, GACGT;## still have 48 attributes left.## After 33 iterations, +20 secs:## rejected 3 attributes: AATG, GAT, GGTT;## still have 45 attributes left.## After 36 iterations, +20 secs:## rejected 3 attributes: TCGC, TGAA, TGAT;## still have 42 attributes left.## After 39 iterations, +20 secs:## confirmed 2 attributes: ATGAT, CGTG;## rejected 3 attributes: ACGT, GGT, GTCGA;## still have 37 attributes left.## After 43 iterations, +20 secs:## rejected 2 attributes: AACGT, TTT;## still have 35 attributes left.## After 49 iterations, +21 secs:## rejected 1 attribute: CAGG;## still have 34 attributes left.## After 51 iterations, +21 secs:## rejected 1 attribute: CAG;## still have 33 attributes left.## After 54 iterations, +21 secs:## rejected 1 attribute: TAGA;## still have 32 attributes left.## After 60 iterations, +21 secs:## confirmed 1 attribute: GAATG;## rejected 1 attribute: GGAG;## still have 30 attributes left.## After 63 iterations, +21 secs:## confirmed 1 attribute: CGTGA;## rejected 1 attribute: GCGC;## still have 28 attributes left.## After 66 iterations, +22 secs:## rejected 1 attribute: GATGA;## still have 27 attributes left.## After 68 iterations, +22 secs:## rejected 1 attribute: ATAG;## still have 26 attributes left.## After 71 iterations, +22 secs:## rejected 1 attribute: ACG;## still have 25 attributes left.## After 74 iterations, +22 secs:## rejected 2 attributes: ATC, GAACG;## still have 23 attributes left.## After 76 iterations, +22 secs:## rejected 1 attribute: GAGT;## still have 22 attributes left.## After 82 iterations, +23 secs:## rejected 1 attribute: TTGG;## still have 21 attributes left.## After 97 iterations, +23 secs:## confirmed 1 attribute: CTG;## still have 20 attributes left.Checking the model outputs
That took a while! Let’s double check the data:
model_df %>%
head()## # A tibble: 6 x 4
## molecule data rf_orig_fit boruta_fit
## <chr> <list> <list> <list>
## 1 ATP <tibble [132 × 1,345]> <S3: randomForest> <S3: Boruta>
## 2 Arginine <tibble [82 × 1,345]> <S3: randomForest> <S3: Boruta>
## 3 Codeine <tibble [94 × 1,345]> <S3: randomForest> <S3: Boruta>
## 4 Dopamine <tibble [108 × 1,345]> <S3: randomForest> <S3: Boruta>
## 5 Elastase <tibble [104 × 1,345]> <S3: randomForest> <S3: Boruta>
## 6 FAD <tibble [92 × 1,345]> <S3: randomForest> <S3: Boruta>model_df %>%
select(rf_orig_fit) %>%
slice(1) %>%
pull()## [[1]]
##
## Call:
## randomForest(x = X, y = y)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 36
##
## OOB estimate of error rate: 8.33%
## Confusion matrix:
## 0 1 class.error
## 0 65 1 0.01515152
## 1 10 56 0.15151515model_df %>%
select(boruta_fit) %>%
slice(1) %>%
pull()## [[1]]
## Boruta performed 99 iterations in 37.49718 secs.
## 35 attributes confirmed important: AAGA, AAGAC, AATT, AATTG, AGA
## and 30 more;
## 1274 attributes confirmed unimportant: AAA, AAAA, AAAAA, AAAAC,
## AAAAG and 1269 more;
## 35 tentative attributes left: AACAC, AACTG, ACACA, AGAC, AGAT and
## 30 more;The output is in the format we expect. The random forest and Boruta models are stored in list columns, and the output looks like that of a random forest and a Boruta model.
The original random forest for ATP does quite well. It has 8.33% out-of-bag error, but misclassifies 10 sequences that contain aptamers are not containing aptamers, which is an issue. Even both Boruta we still might miss some important k-mers.
But Boruta picked out 35 features as important, and left 35 tentative. Reducing over a thousand features to 35 is quite an accomplishment.
Extract features selected by Boruta
Now that we have the initial models, let’s extract the features that Boruta selected and store those
in another list column. Then we’ll append a reduced data.frame with only the features
selected by Boruta (and is_aptamer because we can’t get rid of the response variable!).
Note that map2 is like map, but it takes in two arguments instead of one.
limit_to_selected_attributes <- function(dat, selected_attributes) {
dat_reduced <- dat[, which(names(dat) %in% c("is_aptamer", selected_attributes))]
return(dat_reduced) ## Without the assignment and separate return I get an error with map
}
model_df <- model_df %>%
mutate(selected_attributes = map(boruta_fit, ~getSelectedAttributes(.x, withTentative = FALSE)),
data_reduced = map2(data, selected_attributes, limit_to_selected_attributes))
head(model_df)## # A tibble: 6 x 6
## molecule data rf_orig_fit boruta_fit selected_attrib… data_reduced
## <chr> <list> <list> <list> <list> <list>
## 1 ATP <tibble… <S3: randomF… <S3: Boru… <chr [35]> <tibble [132…
## 2 Arginine <tibble… <S3: randomF… <S3: Boru… <chr [18]> <tibble [82 …
## 3 Codeine <tibble… <S3: randomF… <S3: Boru… <chr [9]> <tibble [94 …
## 4 Dopamine <tibble… <S3: randomF… <S3: Boru… <chr [17]> <tibble [108…
## 5 Elastase <tibble… <S3: randomF… <S3: Boru… <chr [12]> <tibble [104…
## 6 FAD <tibble… <S3: randomF… <S3: Boru… <chr [17]> <tibble [92 …We see that ATP has 35 selected features, Arginine has 18, Coedine has 9, and the others have similar numbers of selected features. As mentioned above, 35 is still fairly large, but it’s much smaller than the 1344 we started with.
Reduced predictor set model fit
The next step is to build a random forest on each reduced predictor set. Then we’ll compare the out-of-bag error between the original predictor set and the reduced set.
set.seed(SEED)
model_df <- model_df %>%
mutate(rf_reduced_fit = map(data_reduced, aptamer_random_forest))Out-of-bag error comparison
Now we compare out-of-bag error for the full predictor set model and reduced predictor set model
for each molecule. We’ll use map_dbl to generate the out-of-bag errors because an out-of-bag error
is of type double. We’ll also calculate the number of features selected by each model for general interest.
Finally, we’ll make a smaller data.frame that only contains the information we need to evaluate the models. This is mostly for readability purposes.
extract_oob_err_rate <- function(rf_model) {
ntrees <- nrow(rf_model$err.rate)
return (rf_model$err.rate[ntrees, "OOB"])
}
model_df <- model_df %>%
mutate(orig_oob = map_dbl(rf_orig_fit, extract_oob_err_rate),
red_oob = map_dbl(rf_reduced_fit, extract_oob_err_rate),
num_selected = map_int(selected_attributes, length))
results_df <- model_df %>%
select(molecule, num_selected, orig_oob, red_oob)
results_df## # A tibble: 23 x 4
## molecule num_selected orig_oob red_oob
## <chr> <int> <dbl> <dbl>
## 1 ATP 35 0.0833 0.0530
## 2 Arginine 18 0.220 0.146
## 3 Codeine 9 0.234 0.170
## 4 Dopamine 17 0.296 0.176
## 5 Elastase 12 0.240 0.144
## 6 FAD 17 0.130 0.0761
## 7 HIV1-Tar 14 0.25 0.163
## 8 Hematoporphyrin 4 0.309 0.206
## 9 IDI4 21 0.173 0.112
## 10 Isoleucine 88 0.0396 0.0432
## # … with 13 more rowsInteresting! It appears that fitting to the Boruta-selected features has resulted in lower out-of-bag error for many molecules. Let’s compare.
Once again, we care about both decrease in out-of-bag error and reducing the number of features to a more managable number. We therefore want to check the following:
What is the median and the IQR of the difference in out-of-bag error across both model types? We look at differences, instead of each model separately, because the difference between “significant” and “not significant” is not itself statistically significant.
What is the median number of features within the reduced models? I’m not looking at the percent decrease because I suspect it will be very high for all molecules. Additionally, if we were to put selected subsequences through standard lab tests, a practical number of subsequences to examine must be a relatively low number, independently of the number of sequences we initially considered.
results_df %>%
summarize(med_orig = median(orig_oob),
med_reduced = median(red_oob),
med_diff = median(orig_oob - red_oob),
iqr_orig = IQR(orig_oob),
iqr_reduced = IQR(red_oob),
med_selected = median(num_selected),
iqr_selected = IQR(num_selected)) %>%
print()## # A tibble: 1 x 7
## med_orig med_reduced med_diff iqr_orig iqr_reduced med_selected
## <dbl> <dbl> <dbl> <dbl> <dbl> <int>
## 1 0.189 0.133 0.0612 0.160 0.0847 18
## # … with 1 more variable: iqr_selected <dbl>set.seed(SEED)
wilcox.test(results_df$orig_oob + rnorm(23, 0, 1e-5), ## Jitter a bit to avoid ties
results_df$red_oob + rnorm(23, 0, 1e-5),
paired = TRUE, conf.int = TRUE, exact = TRUE)##
## Wilcoxon signed rank test
##
## data: results_df$orig_oob + rnorm(23, 0, 1e-05) and results_df$red_oob + rnorm(23, 0, 1e-05)
## V = 263, p-value = 2.098e-05
## alternative hypothesis: true location shift is not equal to 0
## 95 percent confidence interval:
## 0.03591523 0.08653439
## sample estimates:
## (pseudo)median
## 0.06064958We have a median out-of-bag error difference of 6% with an IQR of 16%. The 6% drop is statistically significant according to the result of the paired Wilcoxon signed rank test.
The median number of features selected is 18 with an IQR Of 19.5. That sounds like a reasonable number of molecules to test in a lab setting, especially compared to the 1344 we started with!
Both of these are good signs.
OOB Error Plots
Let’s visualize the difference in OOB-error distributions.
Since we’re interested in the difference in out-of-bag error across both models, we’ll make two plots: one for differences by molecule, and one comparing the distributions of out-of-bag error for both feature sets. We would like to see a drop in out-of-bag error, or at least error that is similar in both models.
Note that we combine plots via the + operator from the patchwork package, which allows
arbitrary plot layouts. Additionally, we use tidyr::gather to combine the two out-of-bag
error columns for boxplots.
diff_oob_plot <- results_df %>%
mutate(error_diff = orig_oob - red_oob,
error_dropped = case_when(error_diff > 0 ~ "Dropped",error_diff < 0 ~ "Increased", TRUE ~ "No Change"),
molecule = fct_reorder(molecule, error_diff)) %>%
ggplot(aes(x = error_diff, y = molecule, group = molecule)) +
geom_point(aes(color = error_dropped), size = 2, show.legend = FALSE) +
geom_vline(xintercept = 0) +
scale_x_continuous(labels = scales::percent) +
theme(panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()) + ## Remove vertical gridlines
ggtitle("Differences in out-of-bag error for random forest k-mer models",
subtitle = 'Positive means "the model on the reduced feature set has lower out-of-bag error"') +
xlab("(Full features model OOB error) - (Reduced features model OOB error)") +
ylab("Molecule")
dists_oob_plot <- results_df %>%
gather(oob_type, oob_val, orig_oob, red_oob) %>%
mutate(oob_type = if_else(oob_type == "orig_oob", "Original", "Reduced")) %>%
ggplot(aes(x = oob_type, y = oob_val)) +
geom_boxplot() +
scale_y_continuous(labels = scales::percent) +
expand_limits(y = 0) +
xlab("Feature set") +
ylab("Out-of-bag error") +
ggtitle("Distribution of\nOut-of-Bag Error\nby Feature Set")
diff_oob_plot +
dists_oob_plot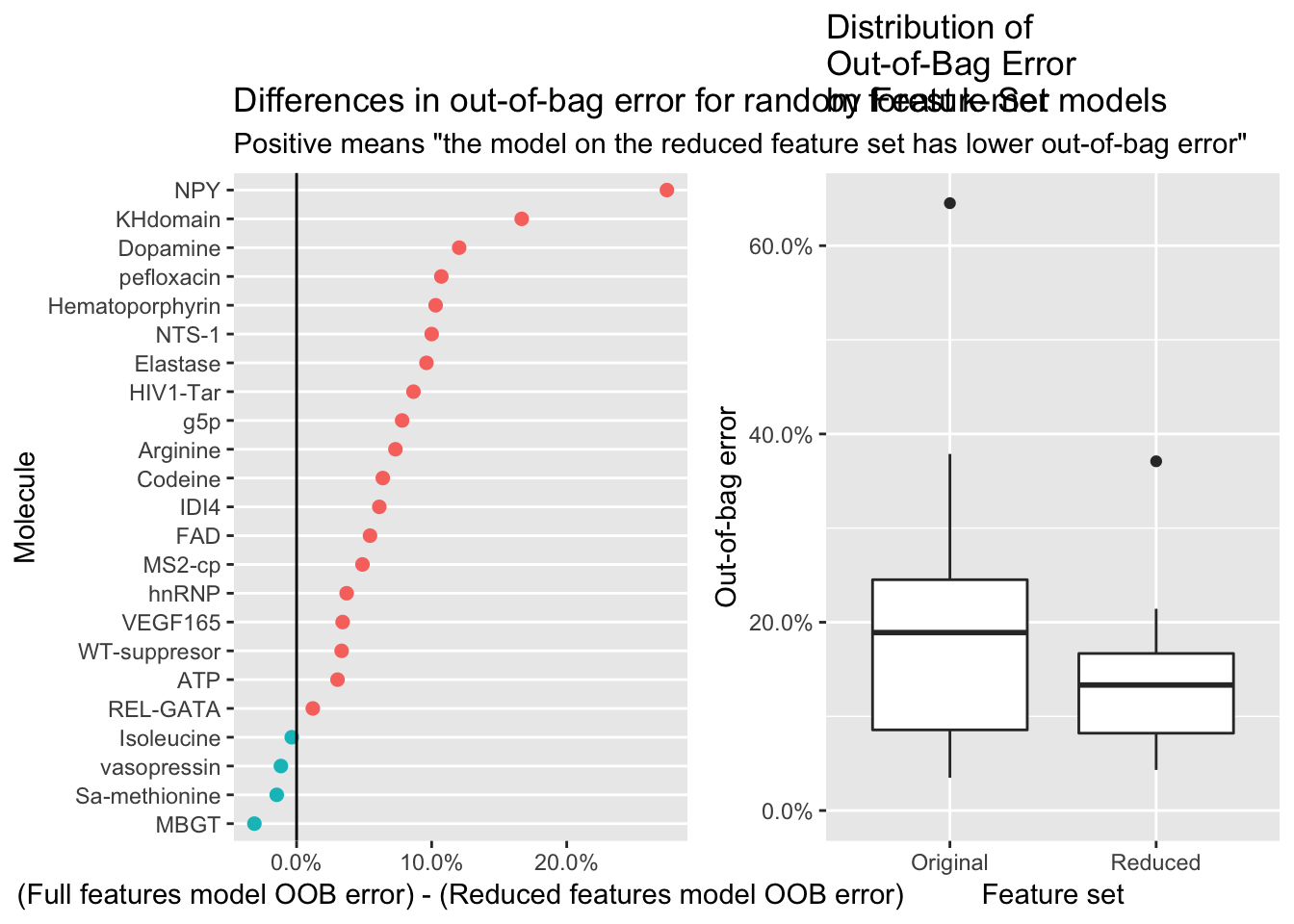
Of the 23 molecules, 19 (82.6%) had decreasing error on the model built on the reduced feature set, and four had increased error. Additionally, the distribution of out-of-bag error is lower overall for the the reduced feature set. This is an indication that in many cases, Boruta does a good job selecting relevant features
Next Steps
In a non-demo situation, biology researchers would take the k-mers selected for each model and would use more definitive, rigorous lab techniques to determine if they are indicative of an aptamer sequence. The authors of the Boruta paper state that the sequences identified by Boruta are consistent with experimental evidence. “…MSA analysis and NMR data suggest that GAAGA motif is responsible for binding adenine in adenine related aptamers. Boruta has found this motif as important in all classes of aptamers binding to three adenine-containing targets that were analysed (adenosine triphosphate, flavin adenine dinucleotide and S-adenosyl methionine).”
If I had more time, I would also have compared the number of features selected to those selected in more typically-seen algorithms, such as LASSO, elastic net, and MARS regression.
Takeaways
I hope that you learned at some of the following from this tutorial:
The complexities of small-\(n\) large-\(p\) problems
How to process data that arrived in a non-standard format
The difficulty of standard feature selection
The power of R’s
tidyverseprogramming environment for data processing
Feel free to contact me on Twitter or LinkedIn if you have any feedback, corrections, or general commentary. Thank you!